From zero to hero: a researcher’s path through data science tools for reproducibility
Leonardo M. Bastos
Assistant Professor
Integrative Precision Agriculture
Slides available at 
Reproducibility?
What is reproducibility?
“Research is reproducible when others can reproduce the results of a scientific study given only the original data, code, and documentation”
This applies to:
- Other people reproducing your work
- Your future-self reproducing your past work
Why bother with reproducible science?
- Tracks how and why of specific decisions and analysis
- Quickly modify analysis and figures
- Increased efficiency
- Rigor and transparency
- Increased citations (of paper, code, script)
But is it really THAT important?
A Nature survey with ~1,600 researchers found that
+70% of researchers have tried and failed to reproduce another scientist’s experiments
+50% have failed to reproduce their own experiments
Main causes: selective reporting, weak stats, code/data unavailability, etc.
My own path on reproducible science: barriers and solutions
🚧 Barrier #1: File naming and management
Starting my M.Sc., this is how my file management system looked like:
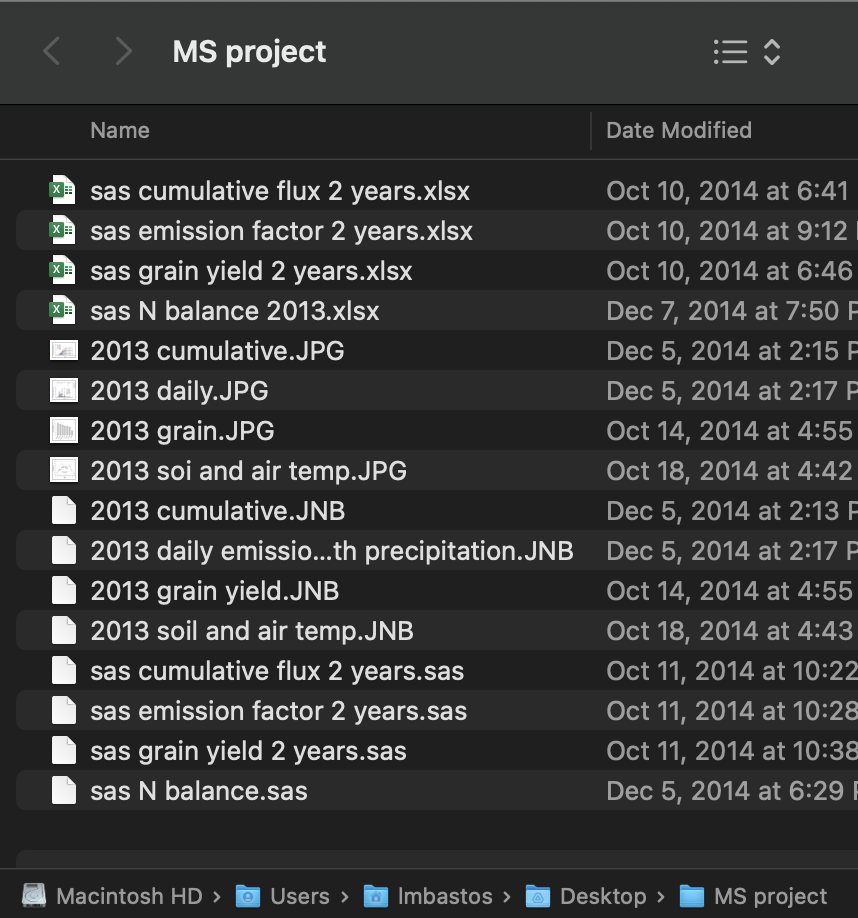
- Data, code, figures all mixed in same folder
- File names not very informative
- And this was just my first year! 😱
- Looks familiar?
🦸 Solution #1: principled file naming and project management
Three principles of file naming
1. Machine readable
- contains key metadata, delimited with “-” and “_”
❌ sas cumulative flux 2 years.xlsx
✅ 2013-2014_N2O-cumulative.csv
- easy to search and filter
- easy to extract metadata
Three principles of file naming
2. Human readable
- name provides info on content, slug concept
❌
daily1.csv
analysisyear1.qmd
figure.png
data2.csv
anova2.qmd
figure2test.png
✅
2013-N2O-daily.csv
2013-N2O-daily-anova.qmd
2013-N2O-daily-plot.png 2014-N2O-daily.csv
2014-N2O-daily-anova.qmd
2014-N2O-daily-plot.png
Which set of files do you want at 3 am before a deadline?
Three principles of file naming
3. Plays well with default ordering
- Start name with something numerical (date, time, experiment number, etc.)
- For dates, use YYYY-MM-DD format
- Left pad numbers with zero
❌
N2O_daily_1-10-2013.csv
10-23-2013_N2O_daily.csv
2-15-2013_N2O_daily.csv
✅
2013-01-10_N2O_daily.csv
2013-02-15_N2O_daily.csv
2013-10-23_N2O_daily.csv
Principles of project management
From this: 
To this:

Principles of project management
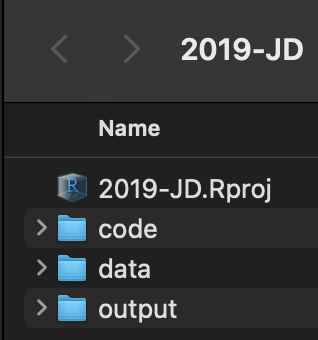
- Create a minimum of three sub-folders: data, code, output
Principles of project management

- Create a minimum of three sub-folders: data, code, output
Keep data files in
data, script files incode, and tables and figures inoutputIn RStudio, use RStudio Projects!
🚧 Barrier #2: Proprietary, point-and-click software
Also during my M.Sc., I was using multiple point-and-click, proprietary software:
- Excel for data organization/manipulation
- SAS for stats
- SigmaPlot for plots
- ArcGIS for maps
Warning
Proprietary software and file extensions hamper reproducibility by imposing a paywall.
Point-and-click is hard to document
Point-and-click software is prone to human error, and normally humans are not good at documenting each step we do.
Think about the last time you organized data in Excel. Do you remember each step you took when filtering or deleting cells, or when creating new columns? What was your decision-making process?
Or maybe each step you took to create a complicated figure. How easy would it be for you to replicate it?
🦸 Solution #2: free programing languages
Free and open-source software
Whenever possible and available, opt for free, open source software.
Instead of this:
- Excel
- SAS
- SigmaPlot
- ArcGIS
Use this:
- csv
- R/Python
- R/Python
- R/Python/QGIS
Code is documentation
plot n2o_ppm
1 101 0.7965260
2 102 1.1163717
3 103 1.7185601
4 104 2.7246234
5 105 0.6050458 plot n2o_ppb
1 102 1116.372
2 103 1718.560
3 104 2724.623Tip
Code is in itself documentation of each step you do. Adding comments with # make it even more understandable.
🚧 Barrier #3: Static programming and environment
Static scripts with .txt or .R
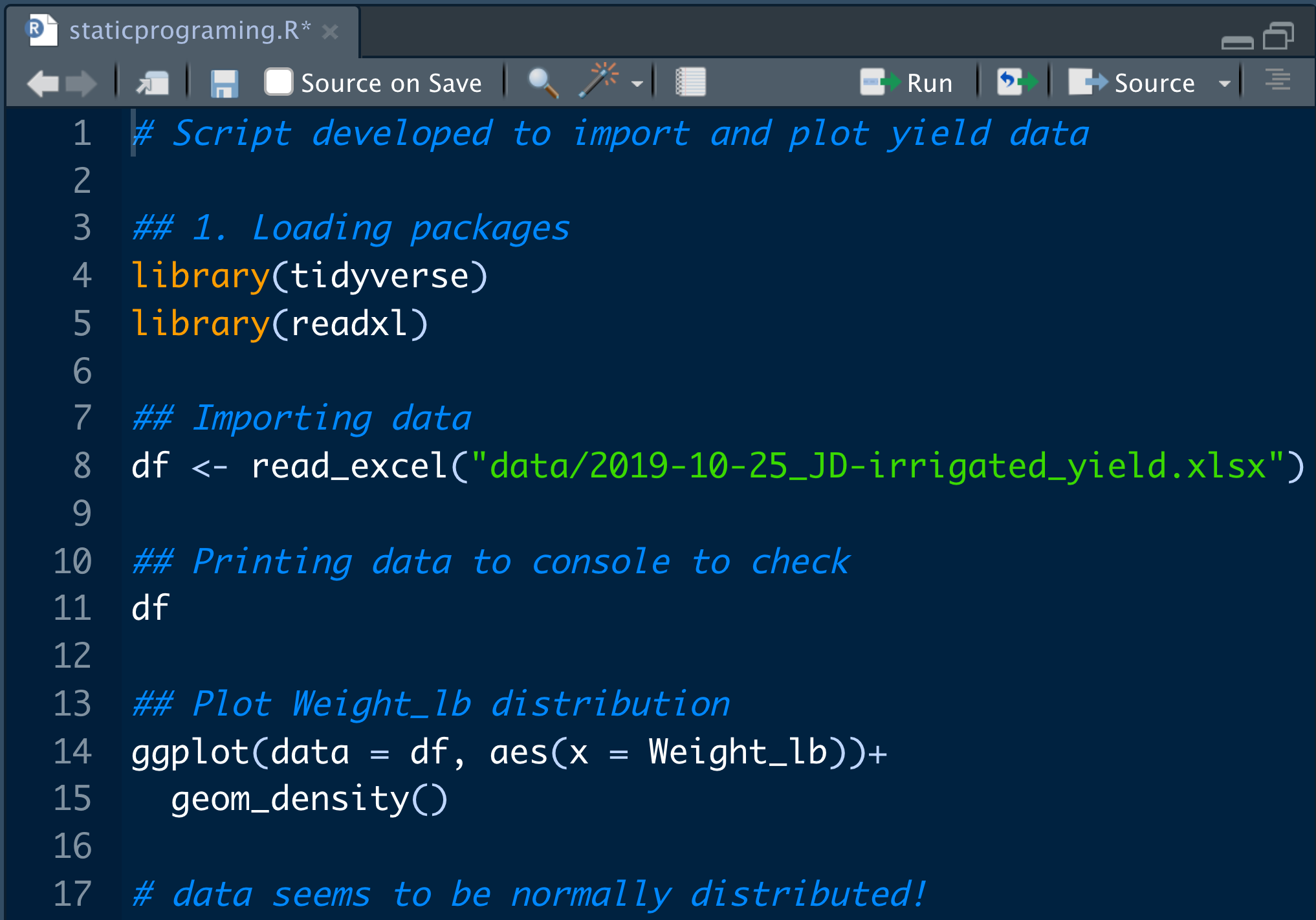
Improvement: using an IDE
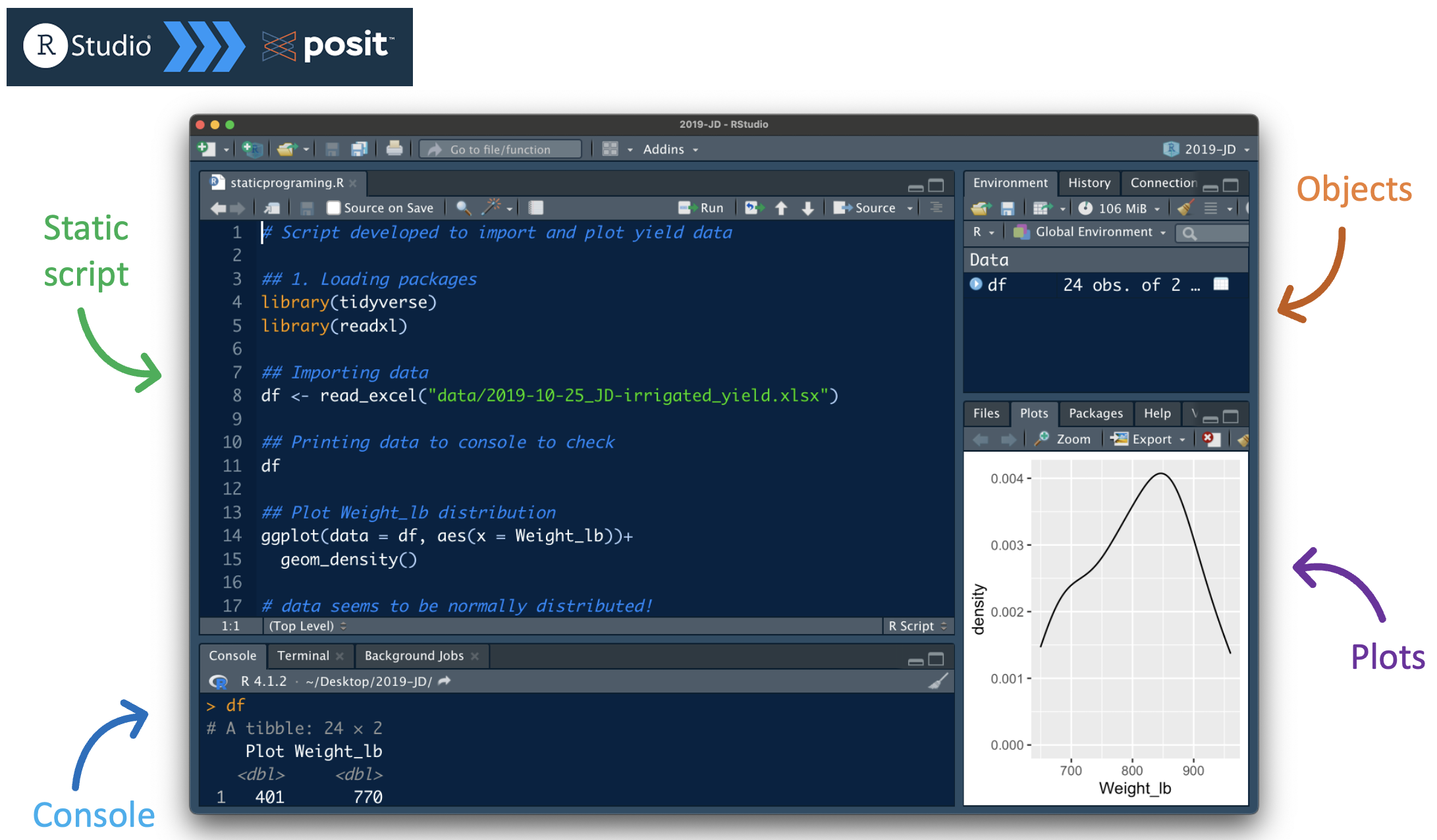
🦸 Solution #3: Literate programming (and IDEs)
Literate progamming
Mixes code, output, and narrative on the same file
Examples:
![]()
quarto + RStudio
🚧 Barrier #4: Keeping track of changes
Can I go back in time?
Situation: you spent the whole week working on an analysis, only to find out it didn’t work as expected OR you got stuck with multiple bugs.
Situation #2: at some point your script had an important piece of code, but at the time you thought you didn’t need it anymore and deleted those lines.
Wouldn’t it be nice/useful/graduate-school-life-saving if you could simply go back in time and start fresh from your latest working version?
🦸 Solution #4: Version control
Welcome in, version control
Think of “track changes”, but on any file type
Especially useful for script files (.Rmd, .qmd)
As your code grows and develops, snapshots are saved allowing you to retrieve different versions
This connects your current-self with your past-self (what were I thinking when I decided on doing this step?)
Locally (in your own machine), use
![]()
🚧 Barrier #5: Reproducibility requires sharing
git works locally
- git is powerful on its own, but it only acts locally
- It becomes really powerful when we can have its features working online
- Working with it online also happens to be perfect for collaboration and sharing 🤝
🦸 Solution #5: Open data and code
Welcome in, GitHub
GitHub is an online centralized platform that combines git, collaborative tools, and cloud storage, all free 💸
You can choose if your projects hosted on GitHub (i.e., a repository) can be seen by everyone (public) or only by you and invited collaborators (private)

GitHub demo
- I’ll show you next one of my GitHub repositories
- This repository was used to conduct the entire analytic flow of a manuscript among 2 collaborators
- Both collaborators had local versions on their computers, and GitHub served as the “merging” point
🚧 Barrier #6: What if software versions change?
Things change
- Computer operating systems get updated
- R gets updated
- RStudio gets updated
- R packages get updated
Even if we are working off of the same GitHub repository, our local software versions may differ, which can cause discrepancies and issues that may impact reproducibility.
🦸 Solution #6: Containerization
Containerizing projects
- To avoid discrepancies of software versions, we can use containers
- Containers keep track of all software versions in a project, and ship that project with those default versions
- This ensures the project is reproducible not only for collaborators, but also your future self
- One example of container software is
![]()
In a nutshell
🚧 ⏩ 🦸 Reproducible science is about…
- Using sensible file names
- Organizing files in sensible sub-folders
- Using free programming language software
- Using literate programming tools
- Using version control locally
- Using distributed version control to collaborate and share data and code
- Using containers
- Others (custom functions, iteration, code peer-review, etc.)
Personal marketing
This entire presentation was made with quarto, and its source code is available on my GitHub
You can find more info on my lab’s website (also made with quarto): Bastos Lab
![]()
You can find my data science teaching material on my blog: agRonomy
Wish to learn and apply these concepts to your own research?
CRSS XXXX: Applications of data science in ag research, Spring 2024

- Thanks! 🙏 💻



